E. coli is a living organism of which the life mechanism is best understood at the molecular level
How does a living organism make use of genes in its genome? The pillar of my research is to unravel its mechanism, and I use E. coli as a model organism.
A genome indicates all genetic information that a living organism has, including genes. DNA is the main body of a gene. DNA is made up of a long sequence of four kinds of substances (bases), and the genetic information is determined by how these four kinds of substances are arranged (base arrangement). Protein is made according to the information, and the protein creates cells and functions in the cells, and carries out life activities.
There are many reasons to make use of E. coli in research. In the first place, E. coli has been used as a model organism to understand the life mechanism at the molecular level for more than 100 years.
E. coli, which is a single-celled organism, is made up of a relatively simple mechanism for a life organism. Its number of genes is about 4,700, which is not so many, while a human, which is a multi-celled organism, is said to have over twenty thousand genes. While a human uses various genes depending on the parts, such as eyes, nose, and ears, and each cell has a different function, E. coli lives with only one cell. In other words, if we investigate one cell, we can observe its entire life. Therefore, it is adequate for the purpose of understanding a living organism.
Moreover, cultivating it is easy and safe. It propagates quickly and can be easily genetically modified. Thus, it has the advantage of being easily handled as a research material.
E. coli is the best understood living organism for various phenomena. In 1997, the whole picture of its genome sequence became clear, and research further diversified. It is also used to produce useful substances such as polymers (high-molecular compound) by altering the metabolism of E. coli. On the other hand, its research as intestinal bacteria, and the research on pathogenic Escherichia coli, such as enterohemorrhagic Escherichia coli O157, and Escherichia coli infection, are also advancing.
The transcription factor controls which gene is used when, where, and how much
I am looking at a protein group called transcription factor (transcriptional regulatory element) in order to understand the mechanism of gene utilization. Which gene is used when, where, and how much depends on its environment and situation. Then, the transcription factor selects the developing gene.
Genetic information of an organism functions by creating protein based on the information on RNA, which is synthesized from DNA. When a living organism tries to develop a certain function, a reaction occurs where an enzyme called RNA polymerase connects with DNA holding such genetic information, and RNA is synthesized using that as a template. This reaction to copy RNA from DNA is called transcription, and the transcription factor connects with DNA while responding to the environmental change, and by interacting with RNA polymerase, it controls the transcription activity.
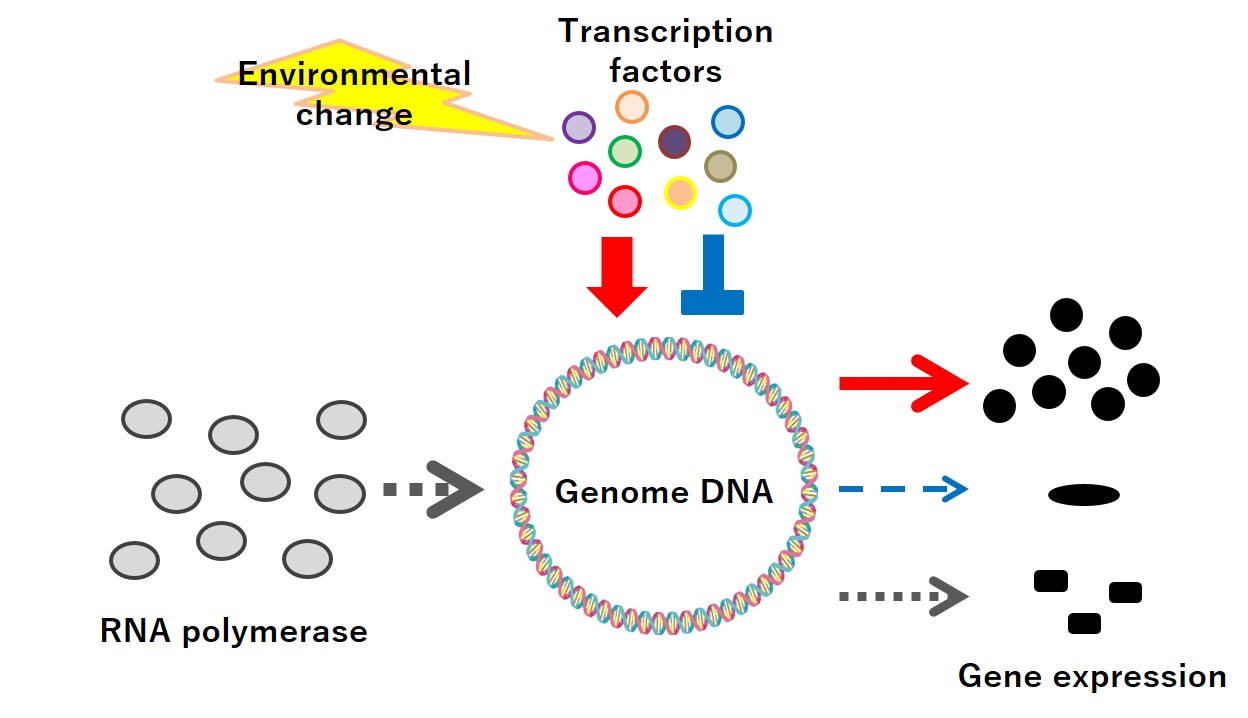
We live in a time when we can tell what kind of gene sequence each organism has and what kind of genetic information it has. If we know that, we are able to estimate what kind of gene the organism has and what it can do. In other words, we can tell what kind of ability that organism has.
However, we cannot tell when that gene is used on what occasion and how much. Some genes are used just once per cell, while some genes are used hundreds and thousands of times. Depending on the change of environment, sometimes a certain gene is decided to be used a lot, and sometimes it is decided to be unused. Using genes that are not necessary is a waste of energy consumption. Life is wise enough to use the necessary energy when necessary, for necessary things.
For example, in the case of E. coli, among about 4,700 genes, about 1,000 genes are used to propagate cells. The rest are used for different purposes, such as responding to the change of environment and maintaining the increased number of cells. They increase in the animal intestine, and when expelled as feces, there is little food. Thus, it uses the ability to maintain rather than the ability to increase. If they go back into the intestine and back in the environment with food, it switches the use of energy for the ability to increase genes.
In the case of E. coli, it has about 300 types of transcription factors while having about 4,700 genes. In the case of microbes, in general, the number is about 6-8% of their number of genes. The more genes there are, the more complicated the adjusting mechanism becomes, and the types of transcription factor increase.
Each individual transcription factor has a different function. It senses the assigned chemical compound and change of environment and changes the parameter so that it can respond to them. By combining these 300 types of transcription factors, it controls which and how much of the 4,700 genes will be used. For example, it could be a function to be exerted when a specific nutrition factor increased, or a function to acquire resistance when it encountered stress such as lowered temperature. As such, it uses a necessary ability selectively according to the change.
A method was developed to identify which transcription factor is adjusting which gene
If we can tell with which part of the genome a certain transcription factor connects, we can tell that it exists for adjusting which gene. We developed the Genomic SELEX method to investigate that. It is a method to identify which specific transcription factor is adjusting which gene and how much. When we mix and let a refined transcription factor and a genome DNA fragment react in a test tube, sometimes that transcription factor connects with one of the genes. If we analyze it, we can identify the controlling network and which gene in the genome a certain transcription factor is controlling.
Twenty years ago, a project to investigate all 300 transcription factors that E. coli has using the Genomic SELEX method started. Currently, I am leading it after having taken it over. At the present stage, we have almost come to understand the relationship concerning the 150-200 transcription factors and about which gene they are adjusting. However, just because it is connected, we cannot determine that it is controlling the relevant genes. We can only determine that by verifying the actual reactions and phenomena, such as having a resistance against certain stress or being related to metabolism. We finished such functional verification with about 70 transcription factors, and we are currently in the process of analyzing them.
If we can identify the functions of 300 types of transcription factors, we can understand the mechanism of the living organism called E. coli, such as what it responds to and which gene it uses according to the change in environment. In other words, we can understand the relation of what an organism cell responds to and which gene it uses to survive.
In the hope that we can apply and develop what we have learned so far, if anything, we are currently working on the research to produce by E. coli biodegradable bioplastics that are degraded in nature. We add the gene that makes plastics in E. coli and let it create an enzyme and a polymer by letting it react in a cell. We know what E. coli eats, what kind of metabolism it makes use of, and what that will be transformed to. Therefore it is easy to introduce the gene and reconstruct the metabolism. In order to create such beneficial substances, together with a company, we are also engaged in research on how we can efficiently let a cell create an enzyme.
Moreover, together with another company, we are conducting joint research to control the survival rate of microbes in the natural environment. Of the CO2 on earth, it is estimated that that emitted from human social activities accounts for about 5%. About 60% is emitted from the breathing of microbes in soil, and the remaining over 30% is from the breathing of microbes in the sea. In this research, we aim to reduce CO2 emissions by enhancing the activity of microbes that are beneficial for plants and obstructing the activity of unnecessary microbes. Currently, by putting E. coli without one of the 300 types of transcription factors in the soil, we are investigating what makes it die earlier and what makes it survive longer.
Furthermore, my ultimate goal as a researcher is to understand the mechanism of the entire cell of a living organism, and to know when and on which occasion the living organism is exerting its ability. If the entire picture of the mechanism of E. coli becomes clear, it will also lead to the understanding of the basic mechanism of living organisms. Some may think that basic research on model organisms cannot directly contribute so much to society. However, by pursuing such research, and if we can understand life phenomena at the genetic level, I think that substantial applications that can be used in a very wide range of areas will be possible.
* The information contained herein is current as of August 2024.
* The contents of articles on Meiji.net are based on the personal ideas and opinions of the author and do not indicate the official opinion of Meiji University.
* I work to achieve SDGs related to the educational and research themes that I am currently engaged in.
Information noted in the articles and videos, such as positions and affiliations, are current at the time of production.


